I am interested in generating and using large-scale datasets for functional inference and hypothesis generation, with emphasis on plant stress biology; validating computationally-derived hypothesis in a biological context, that is, doing “wet lab” molecular work for hypothesis testing; and developing novel data visualization methods and easy-to-use tools for mining data. Some examples of past research are below.
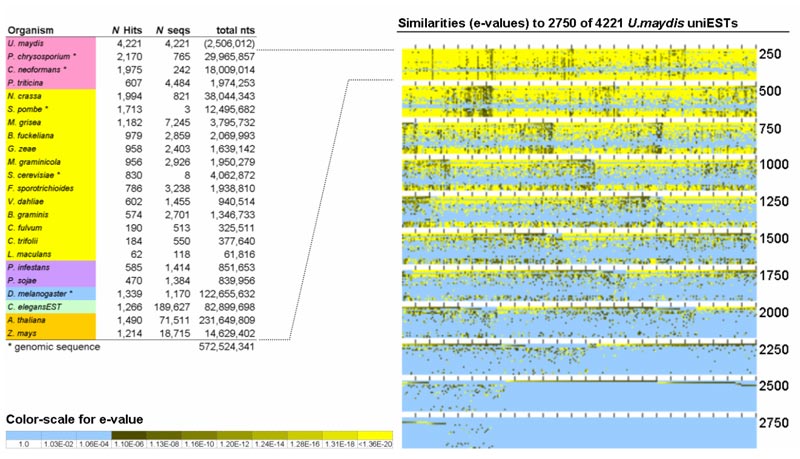
One aspect of my research involves bringing diverse datasets together, and implementing easy-to-use web-based tools with sensible data visualization methods for examining them. Our electronic Fluorescent Pictograph (eFP) browser, for instance, paints gene expression data from various sources onto a pictographic representation of the samples. Shown below is the expression pattern of ABI3, strongest in developing seeds. The eFP Browser and other useful tools are available at the Bio-Analytic Resource, http://bar.utoronto.ca, created and run by my laboratory. We also run the Thalemine aspect of Araport.org and provide data feeds to other international plant bioinformatics portals like TAIR and MaizeGDB.
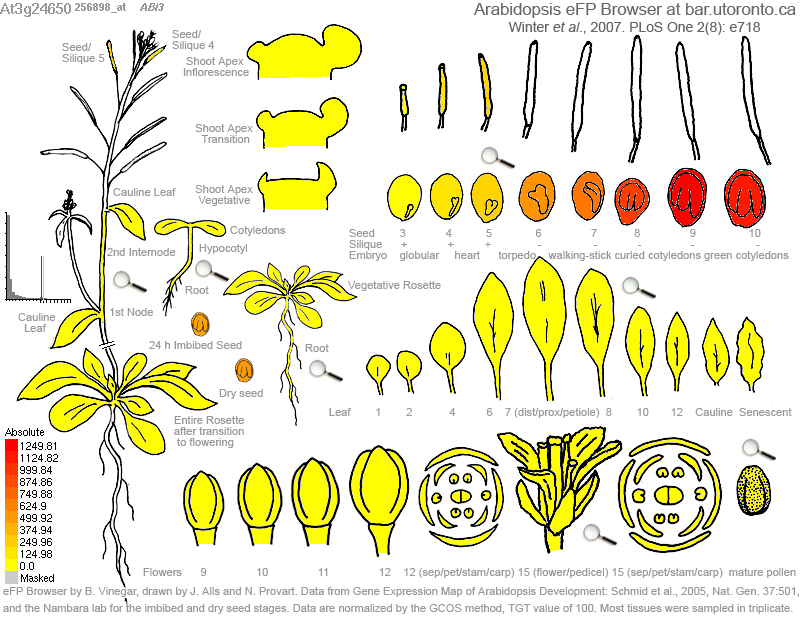
Using gene expression data it is possible to gain insights into plant biology at a system-wide level. For instance, in the following example (Zhu et al. 2003. Plant Biotech J. 1:59-70 [PDF]) the gene expression levels of various enzymes were mapped to the starch biosynthetic pathway of rice over the course of rice grain development. Interesting biological questions are raised by these data: Why are some sucrose synthase isoforms more strongly expressed early in seed development? What is the significance of the shift in transporter expression? Why are different ADP-glucose pyrophosphorylase isoforms expressed in different tissues of the mature grain? Such observations can guide wet lab experiments.
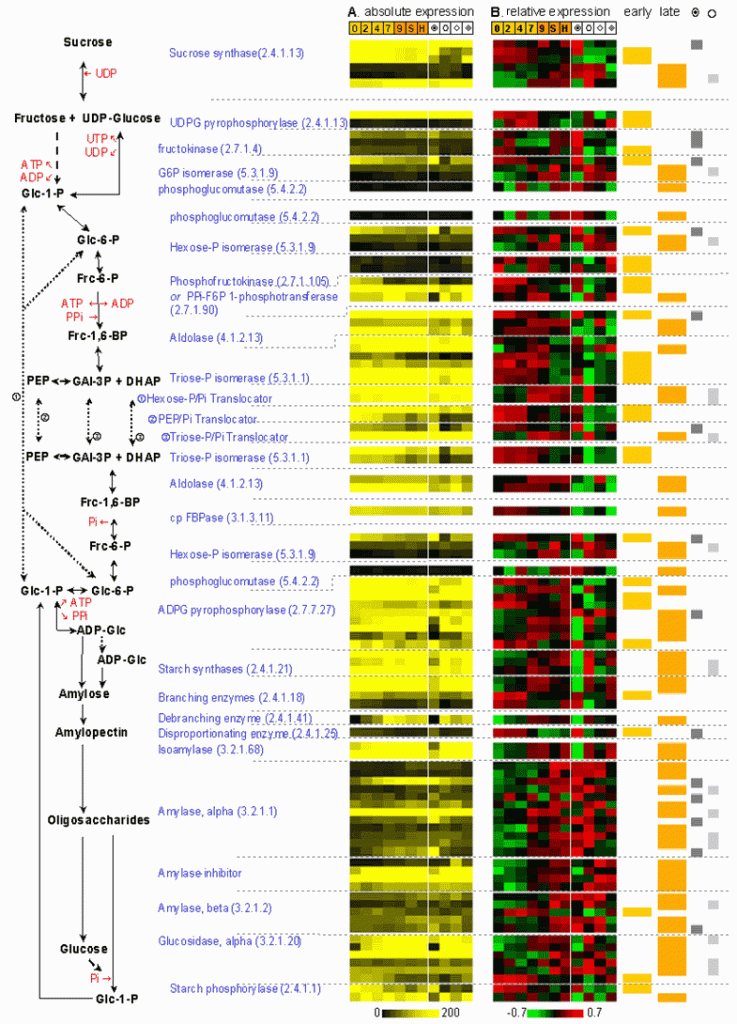
Another possibility for identifying gene function is to use a comparative genomics approach. In this example, a high-throughput BLAST analysis of 4221 ESTs from Ustilago maydis was undertaken by Ryan Austin in collaboration with Barry Saville. The image below shows a subset of the results from almost 100 000 BLAST searches against EST and genomic databases of different plant pathogenic fungi, and of representatives from other organisms. By using such an approach it was possible to identify a small group of genes in Ustilago which are potentially responsible for pathogenesis on corn.